JS Organic Chemistry Learning Objects (JSOCLO) applications
This tutorial is provided to give a high-level overview of the structure and approach used for creating JSOCLO applications. A non-technical reader should be able to get a good sense of how the applications work, while a technical reader should get enough information to understand how to support a JSOCLO application and/or how to approach creating a new one.
Although the applications are written in 100% Javascript, which is a front-end scripting language, conceptually, the applications are implemented using a 3-Tier architecture. The tiers are...
- Data Tier
- Business Logic Tier
- Presentation Tier.
The application structure loosely follows the Model View Controller (MVC) design pattern and uses Delegation as the primary implementation pattern.
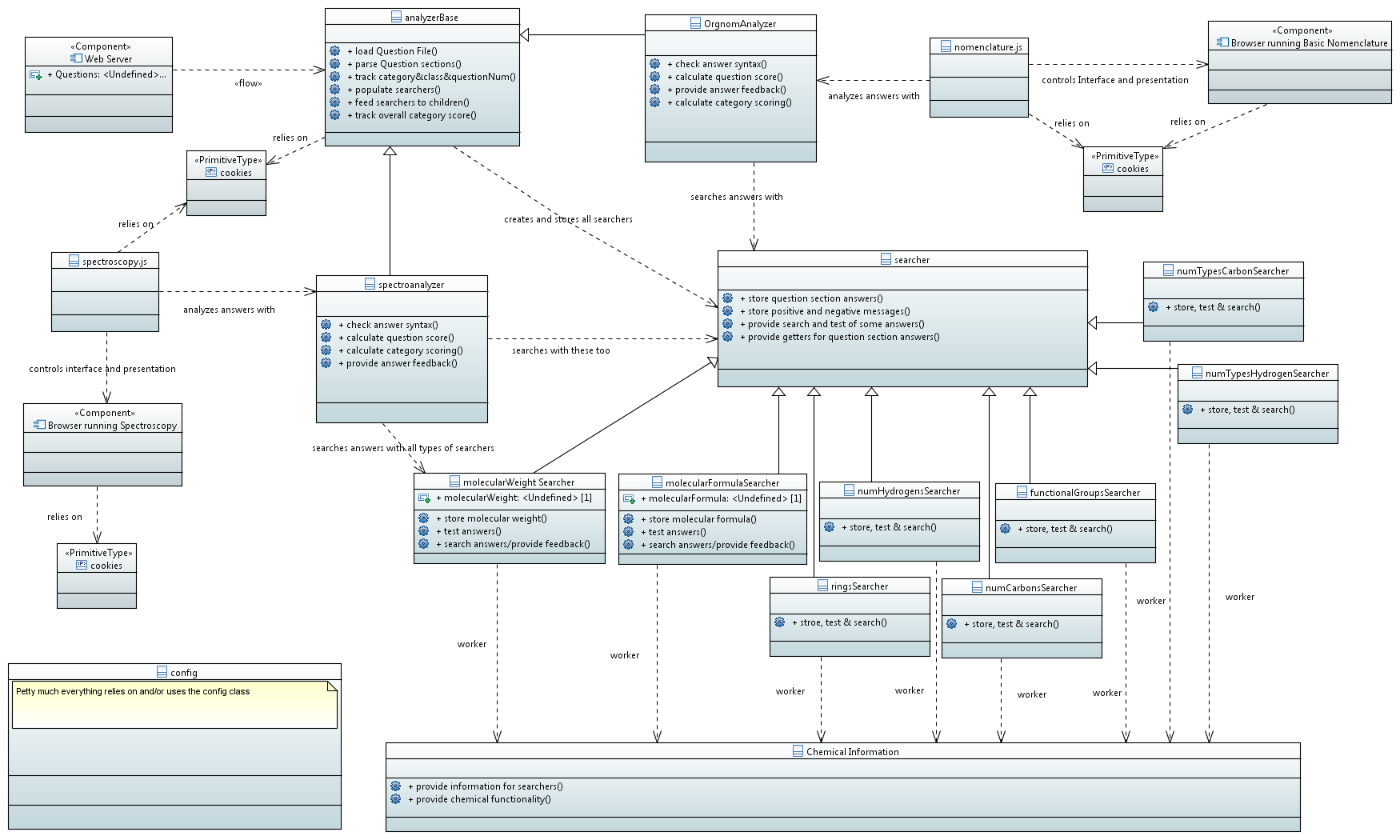
The figure at the end of the document shows the relationships and generalizations of the classes in the applications. It serves to show how any individual application is structured within the larger whole. It also serves to show that each application is structured in the same way. This structure should be used as a model for implementing additional applications to ensure that all applications can be supported in very similar fashions.
Clearly there are differences in presentation, analysis and data access needs between the applications However, structuring all applications such that,
- An application lives in 1 and only 1 html file. (although it may link out to others for help files or support information)
- All presentation and user interface control is within one javascript file.
- All answer analysis is done by 1 application specific analyzer. A child of the analyzerBase class.
- All question file loading and parsing is done by the analyzerBase class.
- Most question file sections are loaded into a searcher class or a child of it. Application specific analyzers can then use these searchers to provide feedback on submitted answers.
For the remainder of this tutorial we'll mostly use Spectroscopy as an example to demonstrate the overall structure for any JSOCLO application .
A note on cookies
JSOCLO takes an in for a penny in for a pound approach towards the use of cookies. If we're is going to use them, then let's use them to their fullest. We do not use any cross domain cookies and we only use cookies to maintain the applications state and scoring. We do not use cookies to track users in any way, shape or fashion. There is simply no need to and no benefit either. JSOCLO applications are intended to provide learning re-enforcement to people who are interested in chemistry, period.
Our use of cookies will not conflict with browser security policies so long as the user has not increased the default levels of security in their browser. The applications have been tested on default installs of the latest(as of July 2014)) IE, Firefox, Chrome and Safari browsers and none have exhibited any issues with our cookies.
A note on JSME
JSME is the Javascript version of JME, both by Brudce Bienfait and Peter Ertl. JSOCLO uses JSME as the graphical portion of the applications. It is used for both presentation of pre-defined molecules and the tool for users to create molecules as answers. In our use we pre-define a div to contain the JSMEApplet(which is not a Java Applet) and use the jsme-nocache.js javascript, which relies on a method called jsmeOnLoad() to create and populate the div. In the html files you'll see a div with id appletContainer; that's the one we use.
jsmeOnLoad() is called by the jsme javascript using a deferred injection. Jsme is a great tool, but it doesn't play nice with others (other javascript libraries). In particular jquery's document.ready function and jsmeOnload() don't play nice together. The setup we use finds the sweet spot for the two. If you see an error like.. "jsme: reference error analyzer not defined...." . the reason is because the document.ready function hasn't been called or isn't finished when the jsmeOnload is called. I'm not sure which one is the case, but the way it is setup now works well.
A note on SMIDGE
Smidge is a lexical analyzer/psuedo parser for SMILES. See
Parser for additional information on smidge. We use it in the
searcher sub classes used in spectroscopy. We parse the submitted
answer SMILE to receive an array of smidge tokens. The searchers pass
the tokens, and sometimes the SMILE as well, to the
chemicalInformation class methods which use the tokens to provide
information back to the searchers; information like the number of
rings or the number of C atoms, or the number of halogens, etc..
A note on the config class
Theconfig class is a static class that provides literals and
constants for all JSOCLO applications. It includes strings used on the
UI, Booleans used to control application behaviour, Error messages used
in various places, URL information for testing and live sites, etc.
Constants of particular importance are (fClass, category,question). analyzerBase and its' children rely on cookies for these 3 constants to be set in order to function. The information is used in the construction of the URL to question files.
In the normal case the presentation layer control javascript (spectroscoy.js) will set these values to some default (based on the config class constant(s)) and then manipulate them as the user interacts. For example, when a new question is selected, spectroscopy.js updates the question cookie and then asks the analyzer to 'reload' or 'loadNewQuestion'.
Construction of the Question file URL:
- fClass: fClass stands for Functional Class and represents an
application specific unique name. In nomenclature, fClass is
'orgnom', in Spectroscopy it is 'spectroscopy'. fClass is used by
various classes and is essential in some cases. Think of it as the
home directory for a specific JSOCLO application.
fClass must always be defined and cannot be the empty string.
- category: category is used to discriminate between questions
on various topics within an application(fClass). In nomenclature, for
example questions are grouped into 'acids','alcohols', 'esters',
etc.. In spectroscopy however all questions fall into a single
category, and as such, spectroscopy uses the empty string for
category in all cases.
category must always be defined but can be the empty string.
As with question numbers, categories are changed by the presentation layers control javascript.
Example, using Nomenclature (nomenclature.js):
function changeCategories(newCategory) { $.cookie("category", newCategory, {path : '/'}); try { . //load question 1 of the new category. analyzer.loadQuestionFile(config.ORGNOM_FCLASS, newCategory, 1); . . - question: question refers to the question number. In the URL
above, examples using values for the question cookie are /question1/question.txt,
/question2/question.txt etc.
question must always be defined and cannot be the empty or a non-numeric string.
Example:
function loadNewQuestion(num) { $.cookie("question", num, {path : '/'}); try { analyzer.loadQuestionFile(null, null, null); . .
The Presentation Tier
The presentation tier for JSOCLO applications is split into 2 distinct layers; literal presentation and presentation manipulation;
Literal Presentation
Literal presentation is accomplished using HTML and CSS. HTML is used to define page elements and to include 3 javascript files. It includes, jQuery so that its document.ready function can be used; jsme because it doesn't play nice if its not loaded via the html and; an fClass control script (spectroscopy.js)
Example:
script src="../js/jquery/jquery-1.11.1.js" type="text/javascript" script src="../js/Jsme/jsme.nocache.js" type="text/javascript" script src="../js/jsoclo/spectroscopy.js" type="text/javascript"
Presentation manipulation
The fClass script provides the second layer in presentation. It is used to;
- load the other requred javascripts, such as the utilities and
analyzers.
Example:function initializeSpectroscopy() { /** * load the other javascripts and CSS */ loadJS("../js/jquery/jquery.cookie.js"); loadJS("../js/jsoclo/chemUtils.js"); loadJS("../js/jsoclo/analyzerBase.js"); loadJS("../js/jsoclo/spectroAnalyzer.js"); loadJS("../js/jsparser/smidge.js"); loadCSS("../css/spectroscopy.css"); - control the elements, such as buttons, inputs, strings, divs,
etc on the page.
Example:// handlers for the buttons. $("#massButton").click(function() { display(config.SPEC_MASS_IMG); }); $("#irButton").click(function() { display(config.SPEC_IR_IMG); }); - manipulate the CSS and html when required. For example,
spectroscoy.js dynamically creates divs with unique names and then
instantiates instances of JSME to populate the div. This is how the
previous answer graphs appear in the feedback section.
Example:// create the jsmeApplet div to display the answer into var jsme = createJsmeDiv(); // insert into the DOM var response = "Your Answer " + jsme.div + "FeedBack: " + feedback; // create the jsmeApplet inside the div var myApplet = new JSApplet.JSME(jsme.id, "250px", "120px", {"options" : "depict"}); //populate the applet with the users answer myApplet.readMolecule(jsmeApplet.jmeFile()); . . - and most importantly act as a bridge to the business logic
tier.
In its bridging capacity spectroscopy.js uses fClass specific analyzers (spectroAnalyzer.js); giving it user submitted answers and requesting feedback to present back to the user; asking if the answer is correct and; telling it to deal with scoring (getCporrect()).
Example:. . //get feedack on the answer var answerFeedback = analyzer.getFeedback(answer); . . if (attempts == config.MAXATTEMPTS && !analyzer.isCorrect(answer)) { unlockQuestion(); . . if (!checkComplete() && analyzer.getCorrect(answer)) { completedQuestions.push($.cookie("question")); . .
The Bussiness Logic Tier
For JSOCLO applications the bussiness logic tier is used to
analyze the submitted answers and provide feedback on them. fClass
specific analyzers are used for this purpopse. For example: OrgnomAnalyzer and spectroAnalyzer. Each fClass analyzer
inherits from the analyzerBase class and then augments itself
with additional methods and members and/or overrides methods in the
base to provide fClass specific business logic.
Shared functionality(shared between fClass applications) remains
in the analyzerBase class and only fClass specific
functionality is added. Examples of shared functionality are common
questionFile sections such as @Correct and @difficulty and/or common
behaviour. some examples:
- All fClass applications share a notion of a correct answer
- All fClass applications could provide hints
- All fClass applications have an fClass, category and question variable.
- All fClass applications share the same notion of question difficluty
- All fClass applications share a notion of questions files that are broken into 'sections' using the @ symbol. See the NomenclatureQuestionFileFormat and SpectroscopyQuestionFileFormat tutorials.
- All fClass applications require their question files to be loaded and the information made availble. (see the data Tier below for more on this)
analyzerBase itself is not intended to be directly instantiated
and includes 2 methods that must be overridden by subclasses and 1 that
may need to be overridden. Those methods are:
- checkSyntax(answer)
- getFeedback(answer)
- isCorrect()
isCorrect() may need to be overridden as well for some fClass applications in the future, but for spectroscopy and nomenclature it is done in the same fashion.
The role of searcher classes in the business tier
It was noted above that all fClass applications share a notion of questions files that are broken into 'sections' using the @ symbol. Conceptually, these sections all do the same thing, they provide something to check against and 1 or 2 responses to give depending on how the check turns out.
For example the "@Correct" section provides the correct answer and a message to present the user when they get the correct answer. The "@difficulty" section provides the difficulty level for the question so that the presentation layer can display the appropriate image. And, the "@search" section provides a regular expression to test answers with and positive and negative feedback messages to provide, based on the test results.
Since the majority of the sections in the question files deal with providing feedback to the user based on the answer they have given, the 'searcher' class was created.
For the most part searchers map to question file sections, and once you have a searcher for a section you don't need another one if that section is reused in another fClass JSOCLO applilcaiton. For example, the @Common section is used in both nomenclature and spectroscopy and both use the same searcher,provided by the analyzerBase to them.
You should think of the question files as being the same for all JSOCLO applicaitons, it is only the sections that may be different between them.
As with analyzers, all searchers inherit from a single base
class.. searcher. This class contains most of the information
that was found in the section, no matter what fClass question file
that section came from. The searcher base class, unlike the
analyzerBase, can be directly instantiated, and is intended for use
with the @search section of any question file.
Once instantiated searcher provides:
- The ability to retrieve either the positive or negative message. (will return the empty string if one or the other was not present in the section).
- The ability to "test" an answer and return a "true" or "false" boolean.
- The ability to "search" an answer and return the appropriate message (based on the return value of the test method).
searcher was used as the base class because all searcher
share commonalities:
- All searchers have either a positive or negative message or both.
- All searchers have something to search against. (regexp, mol weight, chemical formula, etc)
- All searchers share a notion of searching an answer and returning the appropriate message based on the search.
searcher. Testing and
searching however is specific to the kind of section. For example,
searching an answer for its molecular weight is different from
searching for its chemical formula. For this reason, the children must
override both the test and search methods to provide type specific
tests.
For example, below are the complete listing for the molecularWeightSearcher() and the numbrerOfCarbonsSearcher().
/**
* Searcher for Molecular weight.
*
* @class
*
* @extends searcher
* @param {Number}
* mw molecular weight
*
*
*/
function molecularWeightSearcher(mw, correctMsg, wrongMsg) {
searcher.call(this, null, correctMsg, wrongMsg);
var molecularWeight = new Number(mw).valueOf();
var tmp = 0;
/**
* override searcher#test. will return true if the molecular weight of the answer
* is equal to the molecular weight of the question.
* @method
* @access protected
* @param {String} smileAnswer The submitted SMILE string.
*
*/
this.test = function(smileAnswer) {
var tokens = Parser.parse(smileAnswer);
var totalWeight = 0;
var weightMap = chemicalInformation.getMolWeightMap();
for (var i = 0; i < tokens.length; i++) {
var token = tokens[i];
if (token.type == "atom") {
totalWeight += weightMap[token.symbol.toString()];
}
}
// now count the hydrogens
var hydrogens = chemicalInformation.countHydrogens(smileAnswer, tokens);
totalWeight += hydrogens;
if (totalWeight == molecularWeight) {
return true;
}
tmp = totalWeight;
return false;
};
/**
* override searcher#search. will return the approriate message
* from the question file.
* @method
* @access protected
* @param {String} smileAnswer The submitted SMILE string.
*
*/
this.search = function(smileAnswer) {
if (this.test(smileAnswer)) {
if (config.APPEND_CORRECT_VALUES) {
return this.getfoundMsg() + "(" + molecularWeight + ")";
}
return this.getfoundMsg();
}
if (config.PREPEND_INCORRECT_VALUES) {
var me = tmp;
tmp = 0;
return "(" + me + ") " + this.getNotFoundMsg();
}
return this.getNotFoundMsg();
};
this.getMolecularWeight = function() {
return molecularWeight;
};
}
// inherit
molecularWeightSearcher.prototype = new searcher();
molecularWeightSearcher.prototype.constructor = molecularWeightSearcher;
/**
* Searcher for Number of Carbons.
*
* @class
*
* @extends searcher
* @param {Number}
* nm number of carbons
*
*
*/
function numCarbonSearcher(nm, correctMsg, wrongMsg) {
searcher.call(this, null, correctMsg, wrongMsg);
var numCarbon = new Number(nm).valueOf();
var tmp = 0;
/**
* override searcher#test. will return true if the number of Carbons in the answer
* is equal to the number of Carbons in the question.
* @method
* @access protected
* @param {String} smileAnswer The submitted SMILE string.
*
*/
this.test = function(smileAnswer) {
var tokens = Parser.parse(smileAnswer);
var numC = chemicalInformation.countAtomsOf("C", tokens);
if (numC == numCarbon) {
return true;
}
tmp = numC;
return false;
};
/**
* override searcher#search. will return the approriate message
* from the question file.
* @method
* @access protected
* @param {String} smileAnswer The submitted SMILE string.
*
*/
this.search = function(smileAnswer) {
if (this.test(smileAnswer)) {
if (config.APPEND_CORRECT_VALUES) {
return this.getfoundMsg() + "(" + numCarbon + ")";
}
return this.getfoundMsg();
}
if (config.PREPEND_INCORRECT_VALUES) {
var me = tmp;
tmp = 0;
return "(" + me + ") " + this.getNotFoundMsg();
}
};
this.getNumCarbons = function() {
return numCarbon;
};
}
// inherit
numCarbonSearcher.prototype = new searcher();
numCarbonSearcher.prototype.constructor = numCarbonSearcher;
You can see how similar the two classes are. In fact, they could be
abstracted again so that only the test function would have to be
implemented, however, this would make the class harder to initally
understand. test() and search() go hand in hand and leaving them this
way makes it clear what's going on with this searcher, and consequently
its siblings, just by looking at its listing.
analyzerBase is responsible for creating searchers from
question file sections. Children of analyzerBase such as
spectroAnalyzer are responsible for overriding analyzerBase#getFeedback, that is providng feedback to the
presentation layer.
With this in mind, it should become clear that spectroAnalyzer needs
only to use the searchers available to it to construct the feedback
string it returns to the presentation layer. A child analyzers
getFeedback() method is nothing more than the amalgamation of
delegated work.
Example, part of the getFeedback method for
spectroAnalyzer:
...
response += this.checkSyntax(answer);
// there may be more than one common section
// in spectro
var tmp = this.getCommonSearchers();
if (tmp != null) {
var res = "";
for (var i = 0; i < tmp.length; i++) {
var cSearch = tmp[i];
res = cSearch.search(answer);
if(res.length != 0){
response += res;
res = "";
}
}
}
response += this.getMolecularWeightSearcher().search(answer);
response += this.getNumCargonSearcher().search(answer);
response += this.getNumHydrogenSearcher().search(answer);
response += this.getMolecularFormulaSearcher().search(answer);
//these can return empty strings
tmp = this.getRingsSearcher().search(answer);
if(tmp.length != 0){
response += tmp;
}
tmp = this.getStereoSearcher().search(answer);
if(tmp.length != 0){
response += tmp;
}
tmp = this.getChargeSearcher().search(answer);
if(tmp.length != 0){
response += tmp;
}
tmp = this.getFunctionalGroupsSearcher().search(answer);
if(tmp.length != 0){
response += tmp;
}
.
.
getFeedback() delegates the work of getting the feedback to the
searchers made availble to it by its parent class analyzerBase.
The result is a modular structure that makes any particular module fairly easy to understand and an approach that is fairly easy to replicate for future JSOCLO applicaitons while minimizing duplicated code.
The role of the chemicalInformation class
Have a look at the test() methods for the two searcher listing
above. The general approach in the highlighted box above is carried
forward with the searchers test() methods. The searchers test methods
divide to conquer by delegating the chemical related work to methods
of the chemicalInformation static class.
The Data Tier
The analyzerBase class deals with question related data
needs of all the JSOCLO application. As described above it uses the
fClass, category and question cookie values to create a URL to the
question file (question.txt in all cases) for any particular question.
Before instantiating the fClass specific analyzer, spectroscopy.js sets the cookies. It must be done in this order because the analyzerBase uses the values to load the initial quesiton file from the server. Similarily, when categories or questions are changed on the user interface, usually the cookies will be updated and then a call is made to the analyzer to reload itself or to load a new question.
Ajax is used to retrieve the question files from the server. We use Ajax in a synchronous mode (so I guess we should call it Sjax) for 2 reasons. First, on inital page load jsme needs the analyzer to be available to load the intial question. The information it requires is in the question file on the server. When it's loaded asynchronously the jsmeOnload() method will fail.
Second, when a user changes questions or categories there is no benefit in loading the question asynchronously.
The success callback handler for loading a Question is the
parseQuestionFile(data,success) method of the analyzerBase
class. The method splits the file, using @ as the delimiter for
sections, then loops through each section. The loop uses regular
expressions to determine what section is in the current loop iteration
and passes the section to a section specific parser to deal with.
(delegation again).
Example:
function parseQuestionFile(data, status) {
// Split the sections into an Array
var Arr = data.split("@");
try {
for (var int = 0; int < Arr.length; int++) {
section = Arr[int];
// determine the type of section and
// dispatch accordingly
//used in orgnom and spectro
if (/^correct/i.test(section)) {
parseCorrect(section);
continue;
}
//used in orgnom and spectro
if (/^difficulty/i.test(section)) {
parseDifficulty(section);
continue;
}
.
.
In most cases the parse function will split the section into lines and
create a new searcher (using one of the existing ones if possible and a
new one if not) from the information obtained.
Examples:
function parseFunctionalGroups(section){
var lines = getSectionLines(section, /^fg\s*/i);
spectroSearchers.fGroups = new functionalGroupsSearcher(lines[0].trim(), lines[1], lines[2].substr(1));
}
function parseCharge(section){
var lines = getSectionLines(section, /^charge\s*/i);
spectroSearchers.charge = new chargeSearcher(lines[0], lines[1].substr(1));
}
In parseFunctionalGroups, the new searcher is created with two messages (lines[1] and lines[2]substr(1)), being the correct message and the incorrect message (with the $ at the front removed). Lines[0].trim() is the string with the functional group names in it.
In parseCharge, the new searcher is created with only the incorrect message because the @charge section only has an incorrect message associated with it.
The searchers, which are both specific to spectroscopy are stored in an fClass specific data type. The searchers common to both spectroscopy and nomenclature are stored in non specific variables (like, var correctSearcher) of the analyzerBase class.
The spectroSearchers dataType:
var spectroSearchers = {"mw": molecularWeightSearcher,
"stereo": searcher,
"typesC" : numTypesCarbonSearcher,
"typesH": numTypesHydrogenSearcher,
"numC" : numCarbonSearcher ,
"numH" : numHydrogenSearcher,
"charge" : chargeSearcher,
"rings" : ringsSearcher,
"molF" : molecularFormulaSearcher,
"fGroups": functionalGroupsSearcher};
Adding additional sections to be parsed.
Step by step:- Define a unique name for the section (like @MoleculeName ) and use it to add the section to a file.
- Add an if clause to the parseQuestionFile method matching the section with a reqular expression (like /^MoleculeName/i) and use it to delegate the section to a parser for it (like parseMoleculeName(section)).
- Write the parse method (see examples above) and store the new searcher in the class somewhere.
- Write a getter method to retrieve the new searcher.
- Done
Pseudo Class Diagram